Overview
This blog explains techniques for customizing open-source small language models (SLMs) such as Llama 3 and Phi-3 for protein language modeling, enabling users to create novel protein sequences across a broader range of protein families using natural language prompts. The blog also outlines methods for compiling and running these enhanced models on Esperanto’s ET-SoC-1 platform. This work facilitates a comparative study of performance and efficiency between conventional GPUs and Esperanto’s innovative RISC-V based general-purpose machine learning accelerator.
Leveraging Language Models for Protein Engineering
Protein engineering, the process of generating novel protein sequences with desired properties, is vital for industries like drug development and chemical synthesis. Traditional methods rely on introducing random mutations into genes, followed by expression, screening, and reproducing improved variants. While effective, these techniques are labor-intensive, time-consuming, and limited by existing protein templates, making it difficult to create diverse proteins with new capabilities. Additionally, analyzing numerous variants can waste valuable experimental resources.
However, leveraging a language model (LM) that has learned the “protein language” accelerates the process, since it can generate and evaluate protein sequences in seconds. The inherent randomness of LM-generated sequences enhances diversity, enabling the creation of novel proteins with potentially unprecedented functions. This streamlines discovery and expands the possibilities in protein engineering.
Widely Used Protein Language Models
Existing protein language models have been evaluated on a metric which quantifies the stability and order of the generated proteins. This metric is called the predicted local distance difference test (pLDDT). Higher values are better. pLDDT is calculated using sequence-to-structure models such as AlphaFold and ESMFold. Following are the average pLDDT values of some common protein language models.
Current protein language models are based on older LMs that are usually offered as closed source models. As a result there are few, if any, corresponding evaluation datasets that can be used to test the quality of their inferencing results.
| ProtGPT2 | ProGen2 | ESM-3 | ProLLaMA | |
| Developer | Noelia Ferruz Lab | Salesforce | Evolutionary Scale | Peking University |
| Model Size | 774 million | 6.4 billion | 98 billion | 7 billion |
| Open Source | Yes | Yes | No | Yes |
| pLDDT | 56.32 | 61.07 | 80.00 | 66.49 |
Figure 1: Brief Comparison of Widely Used Protein Language Models
Overview of Training/Validation Data Set
Data Sources
- UniProt is the world’s leading high-quality, comprehensive and freely accessible resource of protein sequence and functional information. We have chosen data from SwissProt, the manually annotated and reviewed subset that contains high-quality sequences. Data downloaded from UniProt FTP server.
- Instruction Dataset open sourced by ProLLaMA.
- ECPred40 dataset by Buton et al.
Data Composition
Stage 1: Pre-training
Our dataset contains 2 million protein sequences, and each sequence is converted to the following format:
Seq=<xxx>
Stage 2: Instruction Tuning
Our finetuning dataset includes 10 classes, 780k protein sequences, and the sequences are passed in the following format to pass property along with the sequence:
[Generate yyy protein] Seq=<xxx>
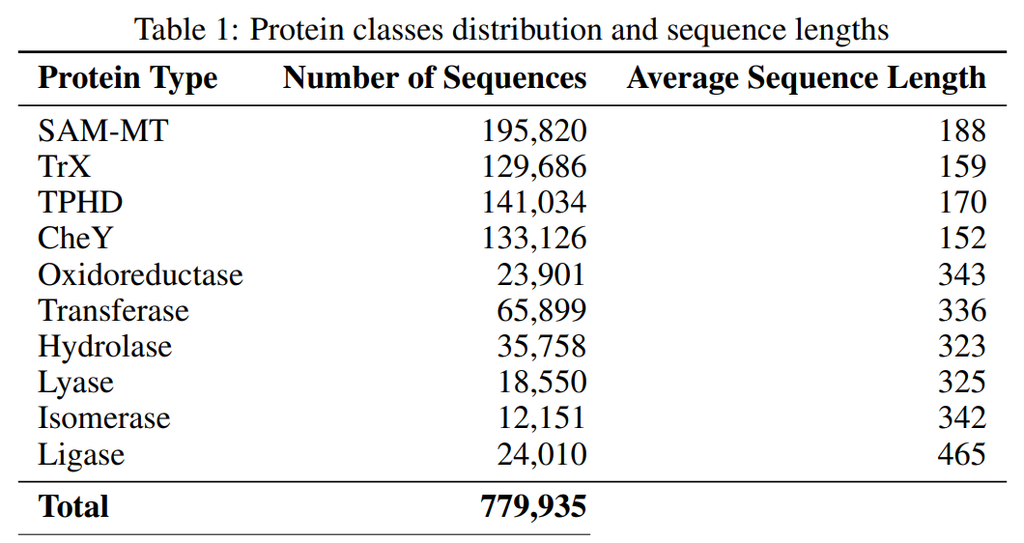
Figure 2: Dataset Introduction [Energy Efficient PLM]
The first four classes shown in the table are for comparison with ProLLaMA as a benchmark. More details can be found here.
The remaining six classes are different types of enzymes that have important roles in essential biochemical processes like metabolism, DNA replication, and cellular signaling: oxidoreductases (catalyze redox reactions); transferases (transfer functional groups); hydrolases (catalyze hydrolysis); lyases (add/remove atoms to/from double bonds); isomerases (rearrange molecules); ligases (join two molecules).
Pipeline of the Project
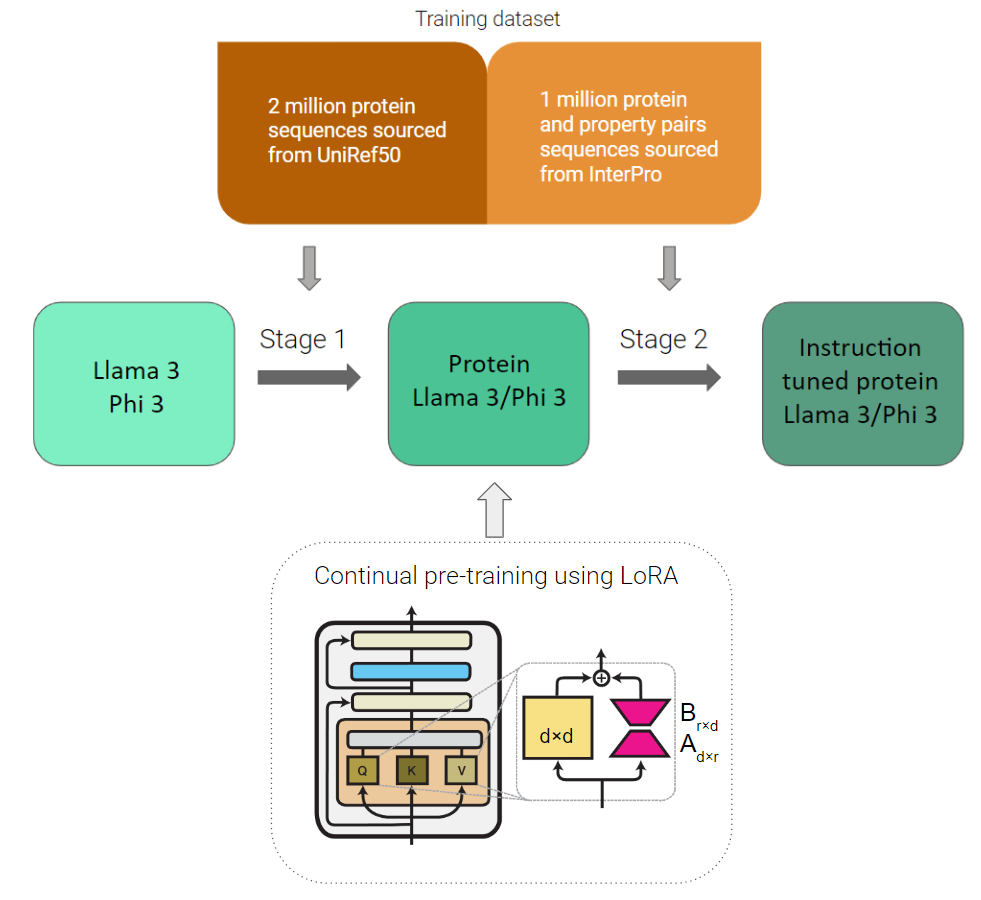
Figure 3: Training Pipeline
Pre-processing
Transform the FASTA data format into the JSON training format.
Model Setup
Download open source model Llama-3-8b and Phi-3-mini-4k-instruct from Hugging Face and enable LoRA adaptors in the attention modules.
For Llama 3:
model = LlamaForCausalLM.from_pretrained(
"meta-llama/Meta-Llama-3-8B",
torch_dtype=torch.float16,
low_cpu_mem_usage=True,
device_map='sequential'
#quantization_config=None
).to(device)
tokenizer = AutoTokenizer.from_pretrained("meta-llama/Meta-Llama-3-8B")
lora_alpha = 256 lora_dropout = 0.05 lora_r = 128 peft_config = LoraConfig(lora_alpha=lora_alpha, lora_dropout=lora_dropout, r = lora_r, bias = 'none', task_type='CAUSAL_LM', target_modules=['q_proj', 'v_proj', 'k_proj', 'o_proj', 'gate_proj', 'up_proj', 'down_proj']) model = get_peft_model(model, peft_config) model.print_trainable_parameters()
For Phi-3:
model = LlamaForCausalLM.from_pretrained(
"microsoft/Phi-3-mini-4k-instruct",
torch_dtype=torch.float16,
low_cpu_mem_usage=True,
device_map='sequential'
#quantization_config=None
).to(device)
tokenizer = AutoTokenizer.from_pretrained("microsoft/Phi-3-mini-4k-instruct", trust_remote_code=True)
lora_alpha = 256 lora_dropout = 0.05 lora_r = 128 peft_config = LoraConfig(lora_alpha=lora_alpha, lora_dropout=lora_dropout, r = lora_r, bias = 'none', task_type='CAUSAL_LM', target_modules=['qkv_proj', 'o_proj', 'gate_up_proj', 'down_proj']) model = get_peft_model(model, peft_config) model.print_trainable_parameters()
Training
Stage 1 – Continual Learning: Instead of pre-training the entire model from scratch on protein sequences, we shall employ the parameter efficient training technique low rank adaptors (LoRA) for facilitating pre-training on protein sequences. This will ensure that the model retains its prior English language understanding along with learning the protein language, hence the term continual learning.
Stage 2 – Instruction Fine-tuning: After the model learns the protein language, it can be further fine-tuned on specific protein classes to help users choose the kind of proteins they want to generate.
training_arguments = TrainingArguments(
output_dir=output_dir,
per_device_train_batch_size=per_device_train_batch_size,
per_device_eval_batch_size=per_device_eval_batch_size,
learning_rate=learning_rate,
gradient_accumulation_steps=gradient_accumulation_steps,
warmup_ratio=warmup_ratio,
fp16=False,
logging_steps=logging_steps,
logging_first_step=True,
num_train_epochs=num_train_epochs,
eval_strategy="steps",
save_strategy="steps",
eval_steps=eval_steps,
save_steps=save_steps,
save_total_limit=save_total_limit,
load_best_model_at_end=True,
report_to = 'wandb',
logging_dir = './logs',
run_name="phi_3_stage_1",
lr_scheduler_type = 'linear',
weight_decay=weight_decay,
log_level = 'debug',
)
hyperparameters = {
"num_train_epochs": training_arguments.num_train_epochs,
"learning_rate": training_arguments.learning_rate,
"per_device_train_batch_size": training_arguments.per_device_train_batch_size,
"per_device_eval_batch_size": training_arguments.per_device_eval_batch_size,
"warmup_ratio": training_arguments.warmup_ratio,
"weight_decay": training_arguments.weight_decay,
"gradient_accumulation_steps": training_arguments.gradient_accumulation_steps,
"max_grad_norm": training_arguments.max_grad_norm,
"lora_alpha": lora_alpha,
"lora_dropout": lora_dropout,
"lora_r": lora_r,
"optim": training_arguments.optim,
"lr_scheduler_type": training_arguments.lr_scheduler_type,
"eval_steps": training_arguments.eval_steps,
"save_steps": training_arguments.save_steps,
"logging_steps": training_arguments.logging_steps,
"eval_freq": training_arguments.eval_steps,
"save_total_limit": training_arguments.save_total_limit,
"load_best_model_at_end": training_arguments.load_best_model_at_end,
"report_to": training_arguments.report_to,
"logging_dir": training_arguments.logging_dir,
"run_name": training_arguments.run_name,
"fp16": training_arguments.fp16,
"eval_strategy": training_arguments.eval_strategy,
"save_strategy": training_arguments.save_strategy,
}
Fine Tuning Configuration
Training Curve
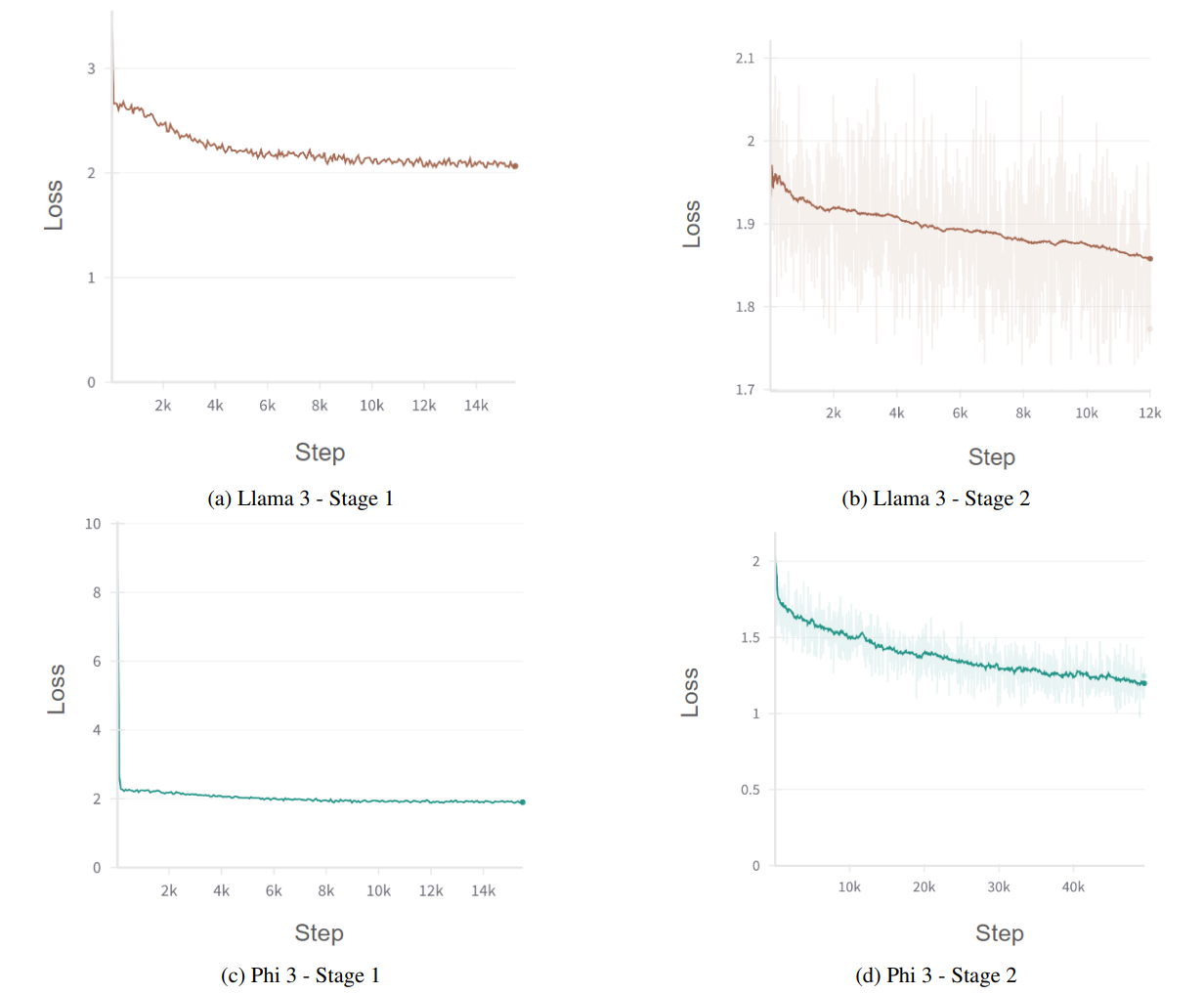
Figure 4: Training Curves for Llama 3 and Phi-3 During Both Stages of Training
User Interface
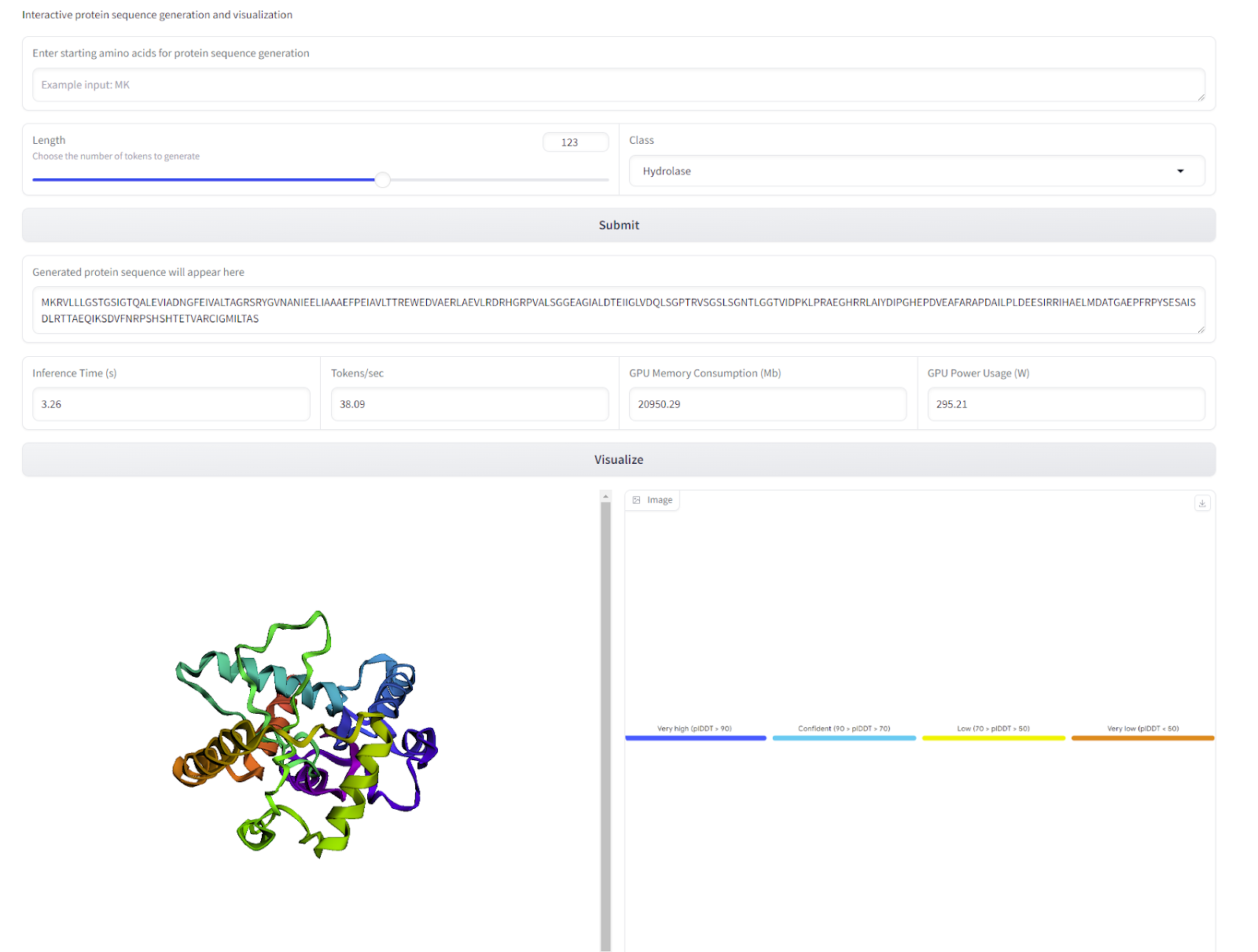
Figure 5: Interface Example
Sample demo space (with limited functionality) can be found here.
The user specifies the class of the protein sequence along with the length and starting amino acids of the protein they want to generate, then presses the submit button. The output will appear in the text box along with important information about the results.
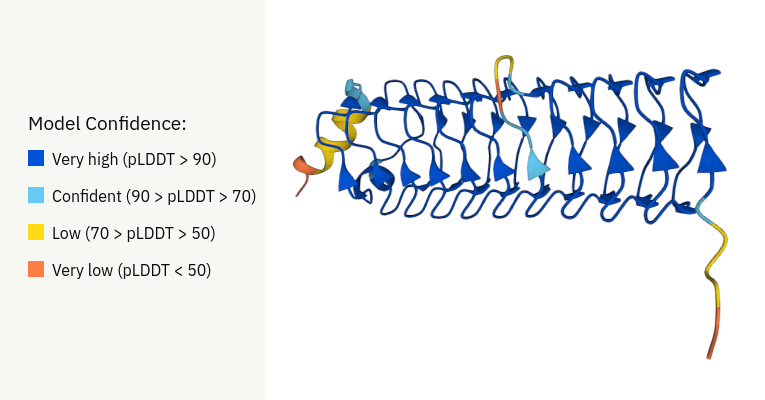
Second, pressing the visualize button will call ESMFold in the backend and predict the structure of this protein, with color coded pLDDT information.
Here is a sample of the video showing the fine-tuned model running on the ET-SoC-1 high efficient platform:
Figure 6: CheY-like Superfamily Protein Sequence Prediction Running on ET-SOC-1 Silicon
Evaluate Effectiveness of Inference Model/Pipeline
Key Metrics
1. pLDDT: Confidence level of the predicted structure.
Stability of generated protein sequences: Stability is a critical aspect of protein sequences, as it determines their potential functionality and usability in various applications, including drug development and industrial processes. The stability is often evaluated using metrics like pLDDT (predicted Local Distance Difference Test), which measures the confidence in the predicted 3D structure of the protein.
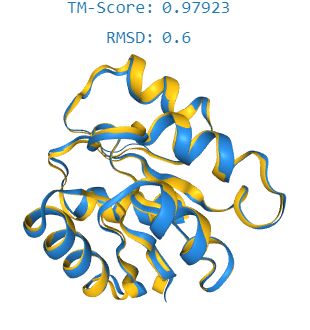
2. TM-Score: Similarity between the predicted structure and a reference structure.
Sequences should exhibit the desired properties: For controllable generation (i.e generating proteins given a user-defined characteristic), it is important to evaluate whether the generated sequences possess the properties mentioned by the user in their prompt. To evaluate this, we use a metric called TM-Score, which is a metric which specifies the alignment of the 3D structure between a given protein and a set of proteins having a common property. The higher the TM-Score, the higher the resemblance between two proteins. By understanding natural language inputs, the proposed method is expected to generate proteins with specific desired properties more effectively than existing models.
Tools and Frameworks
Evaluation Pipeline
After generating 1000 sequences with the fine-tuned model, we calculate pLDDT with ESMFold and TM-Score for each of the properties using Foldseek.
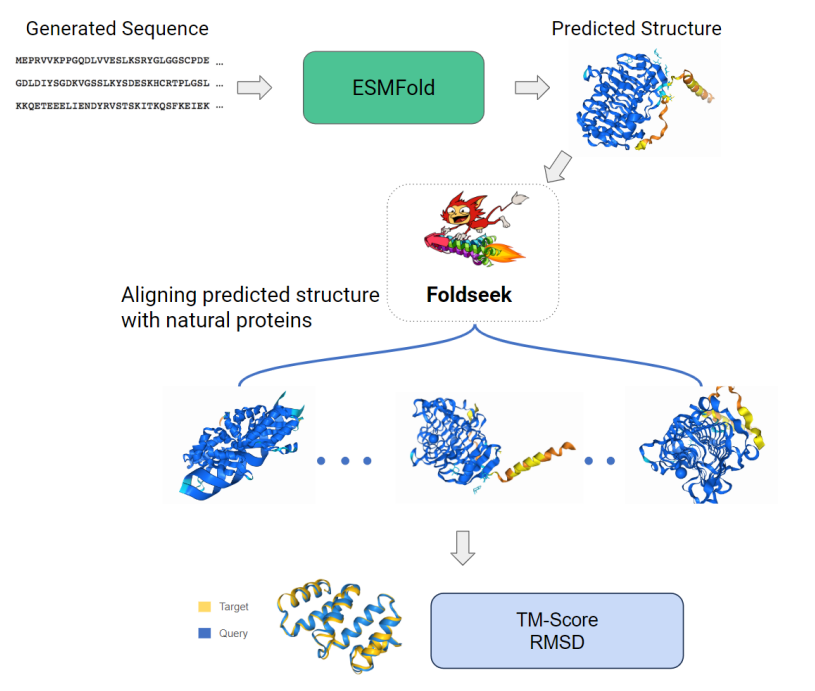
Figure 7: Evaluation Pipeline
Results
Training Details: Advantages Offered by Using Llama 3 and Phi-3 Models
| ProtGPT2 | ProLLaMA | Llama 3 | Phi-3 | |
| Architecture | GPT-2 Large | Llama-2-7b | Llama-3-8b | Phi-3-mini-4k-construct |
| Hidden Size | 1280 | 4096 | 4096 | 3072 |
| No. Attention Heads | 36 | 32 | 32 | 32 |
| No. Training Sequences | 50 million | 50 million | 2 million | 2 million |
| Learning Rate | 0.001 | 0.05 | 0.0005 | 0.0005 |
| GPU Used | 128 A100s | 8 A6000s | 1 A100 | 1 A100 |
| Batch Size Per GPU | 8 | 4 | 8 | 16 |
| Training Time | 4 days | 5 days | 1.5 days | 1 day |
Figure 8: Training Details of Our Llama 3 and Phi-3 vs. Other Protein LM
According to Figure 8, we can see the benefit of our model in reducing the overall training cost incurred. By using a reduced pre-training dataset, we give more focus to instruction tuning and hence reduce the data storage cost by 96%. Our model also converges in less time, with Llama 3 taking 1.5 days and Phi-3 taking 1 day for training respectively. This a 70% reduction and 80% reduction for Llama 3 and Phi-3, respectively, when compared to the 5 days training time of ProLLaMA, proving the utility of our method.
Unconditional Generation (Without Providing the Class of Proteins)
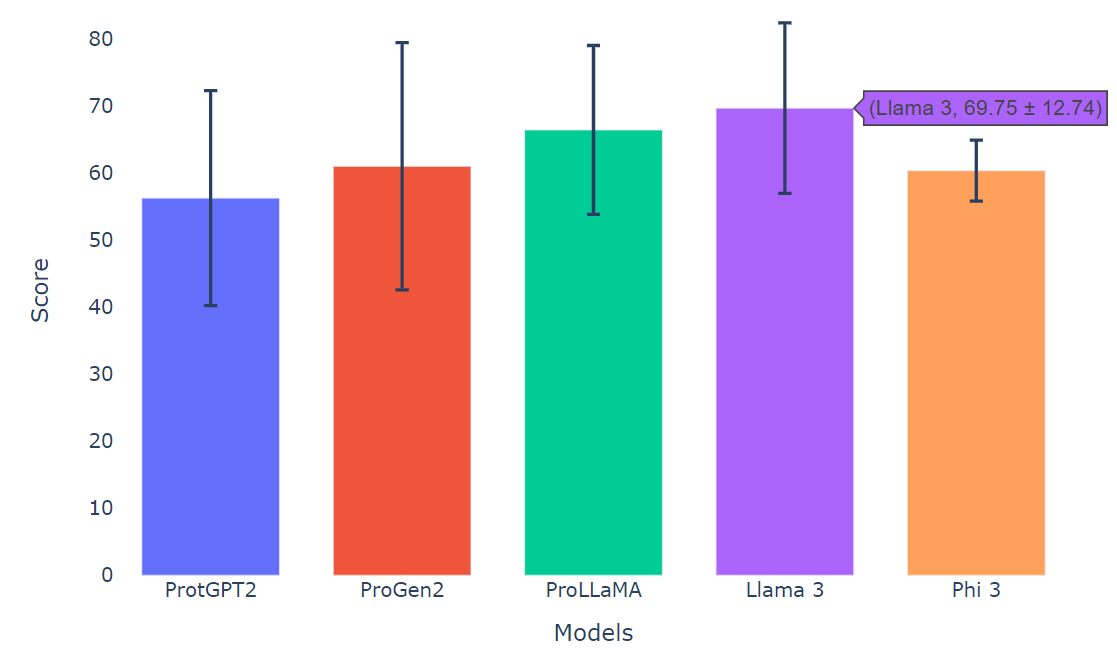
Figure 9: pLDDT Comparison of Various Models
We calculate the pLDDT score across 100 generated sequences to assess model performance. Given the challenges of unconditional generation, our model demonstrates satisfactory results, underscoring the utility of LLM-based methods in de novo protein design. Among the models, Llama 3 achieves the highest pLDDT score, averaging around 69.75 ± 12.74, surpassing ProLLaMa’s score of 66.49 ± 12.61. Additionally, Phi-3 delivers competitive results compared to other models of similar size.
Controllable Generation
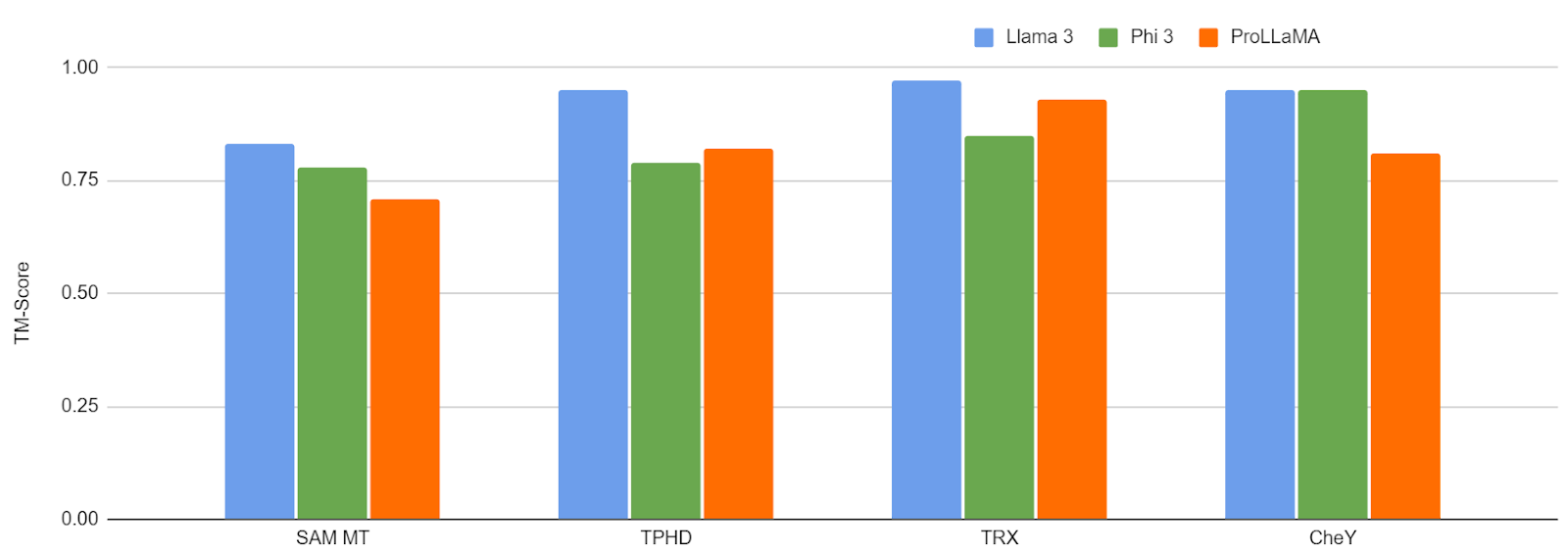
Figure 10: TM-Score Comparison on 4 Properties
Llama 3 consistently outperforms ProLLaMA on the task of controllable protein generation for these four properties. It achieves an average TM-Score of more than 0.8, and hence is able to sufficiently capture the salient features of these classes and generate proteins accordingly. For Phi-3 (3B), the performance is comparable with ProLLaMA (7B).
Comparing Results Between Different Accelerators: GPU and ET-SoC-1
Comparison Criteria
The comparison criteria for evaluating models include several key metrics. Power represents the amount of energy consumed to generate sequences. Tokens per second (Tokens/sec) measures processing speed, indicating how quickly a model can generate or process text. Memory usage provides insights into the hardware requirements and limitations, ensuring models operate smoothly without encountering memory constraints. Finally, transactions per second (TPS) measures the number of transactions completed per second by an information system.
Measured Results
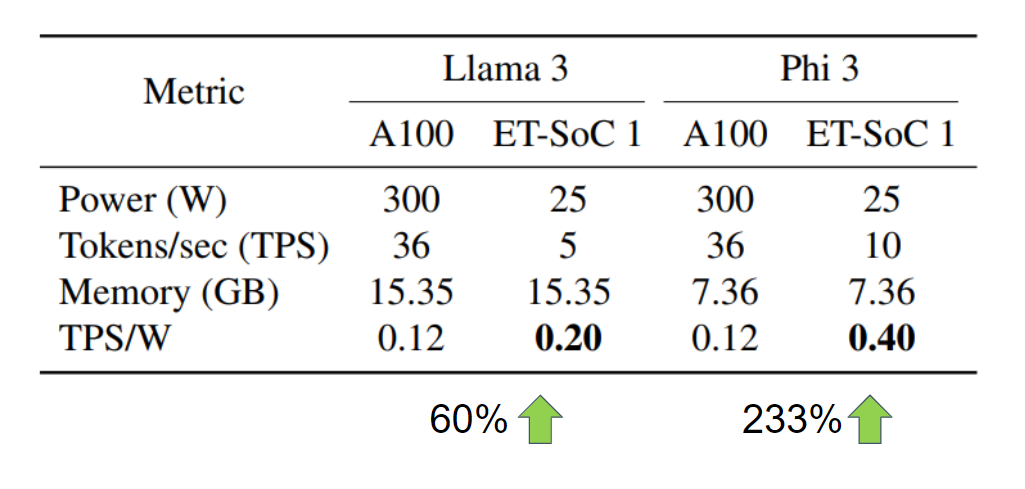
Figure 11: Inference Metrics for GPU vs ET-SoC-1
Comparing inference performance for Llama 3 and Phi-3 when deployed on NVIDIA A100 and ET-SoC-1 in Figure 11, we can see that the throughput, measured in tokens-per-second, decreases for ET-SoC-1. However due to its power reduction, we can see an improvement in throughput-per-watt for ET-SoC-1 by a factor of 3 for Phi-3 and 60% for Llama 3, highlighting the efficiency of the Esperanto platform.
Overall Impact
Primary Issues Addressed
The primary issues addressed in this work include the absence of an evaluation pipeline tailored for a state-of-the-art language model capable of understanding natural language and generating protein sequences. Additionally, we tackle the high overhead associated with fine-tuning.
Contributions
The contributions of this work are as follows: We utilized LoRA to reduce training costs significantly, requiring only 4% of the original trainable parameters. We achieved near state-of-the-art performance with a 96% reduction in the training dataset, using just 2 million data points from the 50 million-point UniRef50 dataset. Additionally, we leveraged latest advances in compact language models to develop two new energy-efficient protein language models based on the LLaMA 3 and Phi-3 architectures. Finally, we expanded the range of protein classes, supporting controllable sequence generation.
Expected Impact
This work has the potential to revolutionize protein sequence generation for industries like drug development and chemical research, where controllable generation is desired. We improved the efficiency and accuracy of generation and decreased training and inference cost by utilizing ET-SoC-1 accelerators.
Future Work
Optimizing Fine-tuning: We can increase the size of the dataset used to include the entire UniRef dataset for further improvement in the results.
Expanding Properties: More properties apart from the 10 used in this work can be included to extend the capabilities of our models.
Structure Prediction: Structure prediction can be improved by fine-tuning ESMFold on our dataset to integrate it into our pipeline for a more unified interface.
Potential Research Collaboration
We would like to extend our congratulations to the 2024 Nobel Prize winners in Chemistry for their groundbreaking work in protein structure prediction. This also shows great potential in utilizing large language models in protein-related areas.
We hope that our work will inspire more scientists and developers to unlock the full potential of large language models in protein sequence generation, and we aim to contribute to the growing body of research that harnesses LLMs for scientific breakthroughs, driving innovation in both theoretical understanding and practical applications.
We are excited to collaborate and extend this pipeline with potential partners interested in a joint development approach. Partners such as molecular biology research labs, drug development companies focused on structure-based or fragment-based drug discovery, and startups developing customized protein language models (PLMs) in search of an efficient inference platform could greatly benefit from this collaboration.
Additional Resources
The models can be found at: Protein Llama 3 and Protein Phi-3.
The code for training and inference can be found at: Llama 3 and Phi-3.
The full technical paper ‘Energy Efficient Protein Language Models: Leveraging Small Language Models with LoRA for Controllable Protein Generation’ can be found at:
References
- Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat Commun 13, 4348 (2022).
- Lv, L., Lin, Z., Li, H., Liu, Y., Cui, J., Chen, C. Y., Yuan, L., & Tian, Y. (2024). ProLLaMA: A Protein Large Language Model for Multi-Task Protein Language Processing.
- Jumper, J. et al. “Highly accurate protein structure prediction with AlphaFold.” Nature, 596, pages 583–589 (2021). DOI: 10.1038/s41586-021-03819-2
- Jessica L. Binder, Joel Berendzen, Amy O. Stevens, Yi He, Jian Wang, Nikolay V. Dokholyan, Tudor I. Oprea
- Shah, A., Guntuboina, C., Farimani, A., PeptideGPT – Generative Design of Peptides using Generative Pre-trained Transformers and Bioinformatic supervision (2024)
- Shah, A, Jayaratnam, S, Energy Efficient Protein Language Models: Leveraging Small Language Models with LoRA for Controllable Protein Generation
